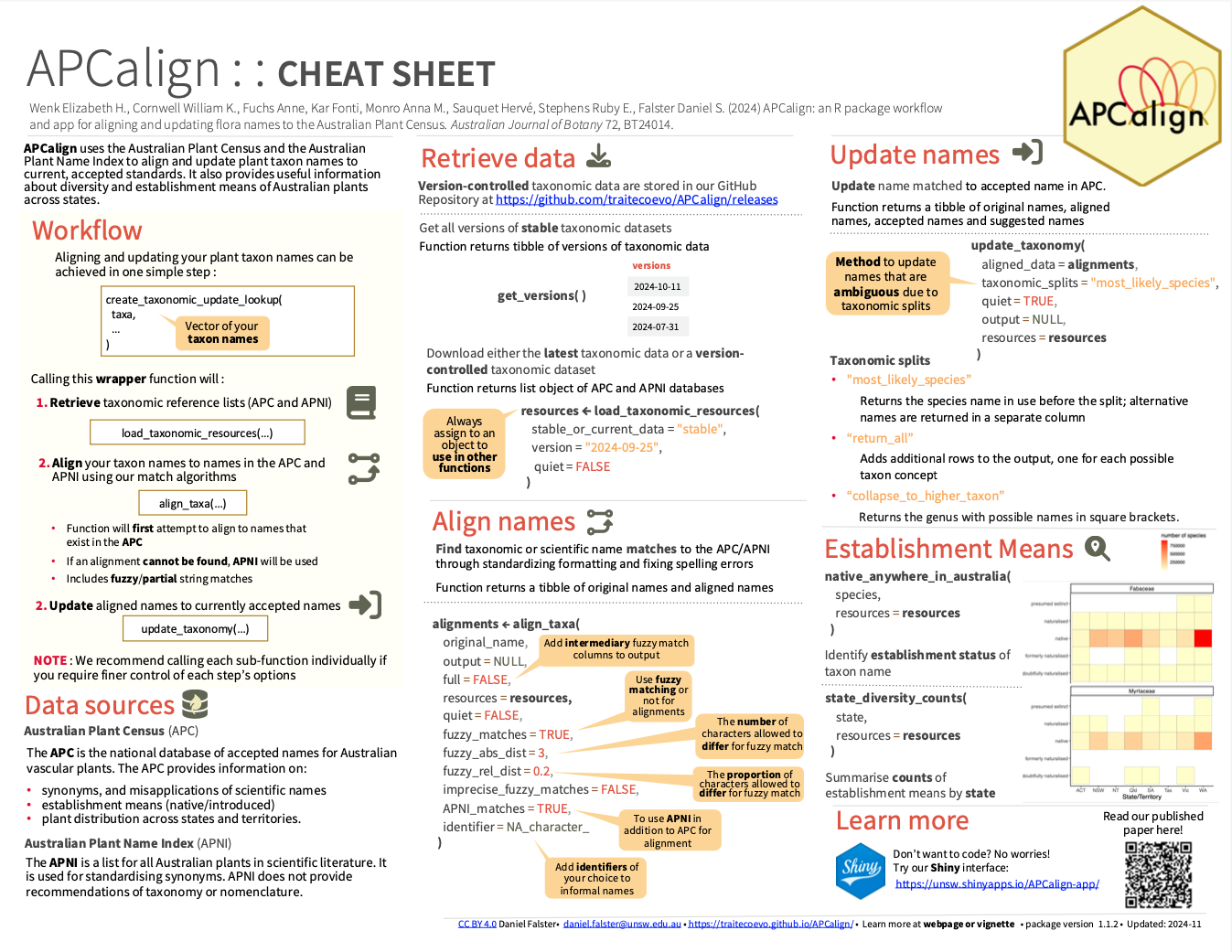
APCalign uses the Australian Plant Census (APC) and Australian Plant Name Index to align and update Australian plant taxon names. ‘APCalign’ also supplies information about the established status (i.e., native/introduced) of plant taxa within different states/territories as compiled by the APC. It’s useful for updating species list and intersecting them with the APC consensus for both taxonomy and establishment status.
DOI: https://doi.org/10.1071/BT24014
Installation 🛠️
From CRAN:
install.packages("APCalign")
library(APCalign)OR for the GitHub version:
install.packages("remotes")
remotes::install_github("traitecoevo/APCalign")A quick demo
Generating a look-up table can be done with just one function:
create_taxonomic_update_lookup(
taxa = c(
"Banksia integrifolia",
"Acacia longifolia",
"Commersonia rosea"
)
)#> ================================================================================================================================================================
#> # A tibble: 3 × 12
#> original_name aligned_name accepted_name suggested_name genus taxon_rank
#> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 Banksia integrifol… Banksia int… Banksia inte… Banksia integ… Bank… species
#> 2 Acacia longifolia Acacia long… Acacia longi… Acacia longif… Acac… species
#> 3 Commersonia rosea Commersonia… Androcalva r… Androcalva ro… Andr… species
#> # ℹ 6 more variables: taxonomic_dataset <chr>, taxonomic_status <chr>,
#> # scientific_name <chr>, aligned_reason <chr>, update_reason <chr>,
#> # number_of_collapsed_taxa <dbl>If you’re going to use APCalign more than once, it will save you time to load the taxonomic resources into memory first:
tax_resources <- load_taxonomic_resources()#> ================================================================================================================================================================
create_taxonomic_update_lookup(
taxa = c(
"Banksia integrifolia",
"Acacia longifolia",
"Commersonia rosea",
"not a species"
),
resources = tax_resources
)
#> # A tibble: 4 × 12
#> original_name aligned_name accepted_name suggested_name genus taxon_rank
#> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 Banksia integrifol… Banksia int… Banksia inte… Banksia integ… Bank… species
#> 2 Acacia longifolia Acacia long… Acacia longi… Acacia longif… Acac… species
#> 3 Commersonia rosea Commersonia… Androcalva r… Androcalva ro… Andr… species
#> 4 not a species <NA> <NA> <NA> <NA> <NA>
#> # ℹ 6 more variables: taxonomic_dataset <chr>, taxonomic_status <chr>,
#> # scientific_name <chr>, aligned_reason <chr>, update_reason <chr>,
#> # number_of_collapsed_taxa <dbl>Checking for a list of species to see if they are classified as Australian natives:
native_anywhere_in_australia(c("Eucalyptus globulus","Pinus radiata"), resources = tax_resources)
#> # A tibble: 2 × 2
#> species native_anywhere_in_aus
#> <chr> <chr>
#> 1 Eucalyptus globulus native
#> 2 Pinus radiata introducedGetting a family lookup table for genera from the specified taxonomy:
get_apc_genus_family_lookup(c("Eucalyptus",
"Pinus",
"Actinotus",
"Banksia",
"Acacia",
"Triodia"),
resources = tax_resources)
#> # A tibble: 6 × 2
#> genus family
#> <chr> <chr>
#> 1 Eucalyptus Myrtaceae
#> 2 Pinus Pinaceae
#> 3 Actinotus Apiaceae
#> 4 Banksia Proteaceae
#> 5 Acacia Fabaceae
#> 6 Triodia PoaceaeLearn more 📚
Highly recommend looking at our Getting Started vignette to learn about how to use APCalign. You can also learn more about our taxa matching algorithm.
Show us support 💛
Please consider citing our work, we are really proud of it!
citation("APCalign")
#> To cite package 'APCalign' in publications use:
#>
#> Wenk E, Cornwell W, Fuchs A, Kar F, Monro A, Sauquet H, Stephens R,
#> Falster D (2024). "APCalign: an R package workflow and app for
#> aligning and updating flora names to the Australian Plant Census."
#> _Australian Journal of Botany_. R package version: 1.0.1,
#> <https://www.biorxiv.org/content/10.1101/2024.02.02.578715v1>.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {APCalign: an R package workflow and app for aligning and updating flora names to the Australian Plant Census},
#> journal = {Australian Journal of Botany},
#> author = {Elizabeth Wenk and Will Cornwell and Ann Fuchs and Fonti Kar and Anna Monro and Herve Sauquet and Ruby Stephens and Daniel Falster},
#> year = {2024},
#> note = {R package version: 1.0.1},
#> url = {https://www.biorxiv.org/content/10.1101/2024.02.02.578715v1},
#> }Found a bug? 🐛
Did you come across an unexpected taxon name change? Elusive error you can’t debug - submit an issue and we will try our best to help.
Comments and contributions
We welcome any comments and contributions to the package, start by submit an issue and we can take it from there!